Wakako Suhara, 1Ichiro Fujimoto, 1,2Katsuhiko Mikoshiba, 1,3Mime
Kobayashi
1,3Touichiro Goto, and 1,3Keiichi Torimitsu
1The Division of Neural Signal Information, NTT-IMSUT
2The Institute of Medical Science, University of Tokyo, 3Materials Science
Laboratory
Mikoshiba’s group identified an abundant glycoprotein that
exists in Purkinje cells of cerebellum as an inositol 1,4,5-trisphosphate
receptor (IP3R) in 1989. IP3R has shown to play pivotal
roles in neuronal transmission and other functions that relate to morphological
and physiological processes in living organisms. Those functions include fertilization,
activation of an egg, axon extension of a neuron, and cerebral nerve plasticity.
Previous analysis of numerous negatively stained images of fixed samples
has indicated that the receptor changes its morphology depending on intracellular
Ca2+ concentrations. The receptor was square-shape in un-liganded
state and windmill-shape in the presence of Ca2+. In this study,
we attempted to observe unfixed samples of a single IP3R protein
by atomic force microscopy (AFM).
The same samples used for the previous electron microscopic studies were
used. Single IP3R tetramers on mica were successfully observed
in imaging buffer without detergent (Fig. 1A). The square-like structure with
a width of about 25 nm closely resembles the model proposed in previous studies.
We expect that individual IP3R molecules assemble in a certain
direction resulting from the electrostatic interaction between IP3R
and the mica surface. After imaging IP3R in standard imaging buffer
without Ca2+, buffer was changed to the one with Ca2+.
We found several types of structures in the presence of Ca2+. Most
notably, we found IP3R tetramer with a dent in the center (Fig. 1C). While
the height decreased slightly, the width clearly increased. The structure
resembles the windmill-like configuration reported in a previous electron
microscope study. The IP3R appeared to be inclining slightly backwards,
and two wing-like structures that were higher than others could be distinguished.
Section analysis along these two higher structures showed a width of 30 nm.
We believe that our study is an important stepping-stone for the application
of AFM to biological samples. Further improvements in AFM will make it possible
to reveal the structural changes of other membrane proteins in a natural environment.
[1] W. Suhara et al., 34th annual meeting of Society
for Neuroscience, San Diego, USA (2004)
[2] I. Fujimoto et al., 9th Linz winter workshop, Linz, Austria
(2005).
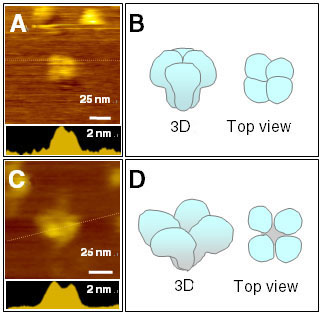 |
Fig. 1. AFM topographs of IP3R. Single
IP3R imaged without (A) or with (C) Ca2+ in
buffer. Section analyses along dotted lines are shown in bottom insets.
Three-dimensional reconstructed models are shown in (B) and (D). |