High-density Functional Protein Arrays
Chandra S. Ramanujan1, Koji Sumitomo2, Maurits R. R. de Planque1, Hiroki Hibino2,
Keiichi Torimitsu2, and John F. Ryan1
1University of Oxford, 2Materials Science
Laboratory
DNA arrays have played a critical role in developing our understanding
of genomics. However, whilst they can measure the expression levels of
large numbers of genes simultaneously, they cannot be used to further characterize
the protein products of such genes and their activity. An important objective
of current research is to extend the use of array technology to both directly
study the function of the proteins to aid drug discovery. Whereas DNA microarray
technology is well-developed, the protein equivalent is still at the early
stages of development. Protein microarrays have been recognized as a valuable
tool since they require only a nanolitre-scale sample volume with a few
picograms of the target protein or drug.
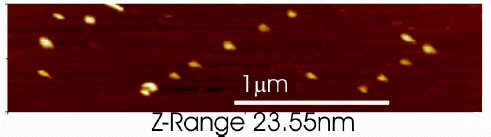
We show that 100 nm unilamellar thiol-tagged vesicles bind discretely
and specifically to Au nanodots formed on a Si surface (See Fig.1). An
array of such dots, consisting of 20 nm Au-Si three-dimensional islands,
is formed by self-assembly on terraces of small-angle-miscut Si(111) after
Au deposition [1]. Consequently, both the formation of the nano-pattern
as well as the subsequent attachment of the vesicles are self-organized
and occur without the need for any ‘top-down’ lithographic processes.
This approach has the potential to provide the basis of a low-cost, high-density
nanoarray for use in proteomics and drug discovery.
Today’s DNA microarray technology has the potential of screening up
to 105 ~106
probes in a 300 ml solution volume. In our nanodot arrays about 109 nanodots
are covered by a 10 ml droplet [2]. The next steps towards producing a protein chip will
include reconstituting proteins into the vesicles and determining a reliable
way of labeling and reading the array, possibly using scanned probe techniques
such as AFM or a combination of AFM and Scanning Near-field Optical Microscopy
(SNOM). The AFM-SNOM has a resolution of about 100 nm. The gold-silicon
combination provides an ideal substrate with a low background for fluorescence
detection techniques. One way of producing a protein nanoarray would be to
first scan and locate each nanodot, and then repeatedly expose to different
protein-vesicle solutions and re-scan, until the chip is fully loaded. The
deposition of a number of different protein-vesicles can be achieved by
optimizing exposure time and vesicle density. The interaction of these mapped
vesicle-proteins with fluorescent-labeled antibodies could be detected using
the AFM-SNOM.
[1] H. Hibino and Y. Watanabe, Surf. Sci. 588 (2005) L233.
[2] C. S. Ramanujan, et al., App. Phys. Lett. 90 (2007) 033901.
 |
||
|