Materials Science Laboratory
The development of biofunctional devices requires a biointerface for maintaining
biomolecules on a substrate without interfering with their structure or
biological functions. By using an artificial membrane biointerface, we
can control the position, density and arrangement of biomolecules, leading
to the design of various bio devices such as highly sensitive biosensors
or implant-type biochips. Here we produced artificial cell membranes on
a substrate that enable proteins to be adsorbed or transported to a desired
position, and thus realized a novel molecular manipulation technique for
use at the biointerfaces [1].
A cell membrane is essentially composed of phospholipids (lipids). The
fluidity of an artificial cell membrane is controlled by the choice of
lipid components. We selectively fabricated artificial cell membranes with
high/low fluidity that contained nickel-chelating lipids (Ni-lipid). The
Ni-lipid is capable of specific binding with terminal modified proteins.
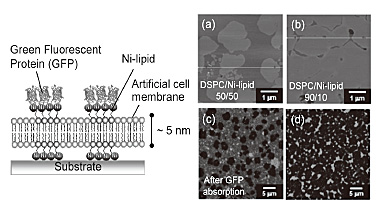
Figure 1(a) and (b) shows AFM images of an artificial cell membrane consisting
of DSPC (low fluid lipid) and Ni-lipid. The image of a 50/50 mol% of DSPC/Ni-lipid
reveals a hetero-geneous membrane structure with round DSPC-rich domains
distributed in a Ni-lipid-rich fluid-phase domain. The DSPC domains were
slightly thicker (1.0 nm) than the surrounding Ni-lipid domain. The area
of each domain was controlled by controlling the molar ratio of the lipid
components. When histidine-tagged green fluorescent proteins (His-tagged
GFPs) were added to the membrane, the fluorescent pattern was evidently
based on the specific adsorption of His-GFPs only in the Ni-lipid domain
[Fig. 1 (c) and (d)]. While the membrane produced by DOPC (high fluid lipid)
and Ni-lipid formed a homogeneous structure with high fluidity. The membrane
exhibited a self-spreading ability in buffer solution whereby a single
lipid membrane (5 nm high) grows on a substrate surface by self-organization
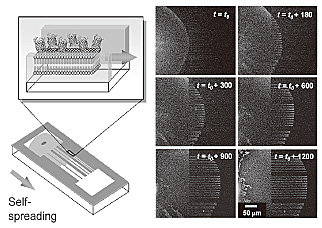
[2]. By employing the self-spreading nature, we demonstrated protein transport
along the micro-pattern. Figure 2 shows the time evolution of the molecular
transport of His-GFPs tethered on the membrane. The transport property
agreed well with the existing self-spreading model of an artificial membrane
[velocity (ν) = (β/time (t ))1/2] where the kinetic spreading coefficient (β) was 10.4 µm2/s.
The biointerface produced by lipids can also employ a cell membrane model to analyze biomolecular interactions or signal transmission events. Therefore, the biointerface will be beneficial for bioscience as well as providing a framework for building biofunctional devices.
[1] H. Nakashima et al., Langmuir 26 (2010) 12716.
[2] K. Furukawa et al., Lab Chip 6 (2006) 1001.
 |
 |
|||||
|
|