John F. Ryan*, and Keiichi Torimitsu
Materials Science Laboratory, *University of Oxford
Receptor proteins play a fundamental role in biological membranes. They
bind to ligand molecules and transfer signals into the cells by means of
electricity and by chemical modification of proteins. Biological membranes
are principally made of lipid molecules. Recent studies have revealed that
lipid membrane in the synaptic junctions of central nervous system (CNS)
is heterogeneous in composition and consists of small regions known as
rafts [1]. These raft-like domains are loci for insertion of receptor proteins
and are believed to act as signalling platforms that organize and compartmentalize
receptor proteins to modify the synaptic signalling.
We report atomic force microscopy (AFM) measurements of the glutamate
receptors (GluRs), the most common membrane receptor proteins found in
the CNS. GluRs are implicated in learning and memory, and up-regulation
of its numbers in the post-synaptic membrane possibly being a key component
of this process. In this work we have investigated the dependence of protein
reconstitution in model membranes on lipid composition.
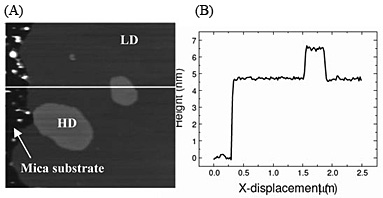
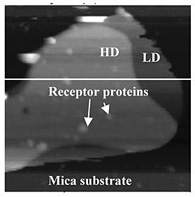
We reconstituted purified GluRs into a lipid mixture system consisting of a mixture that makes up approximately 60% of the synaptic membrane. As shown in Fig. 1, the mixture formed two distinct domains when immobilized on mica: a high domain (HD) at 7 nm height and a low domain (LD) at 5 nm. The lateral extent of HD is typically 〜100 nm, which has structural similarities with the raft-like domains in synaptic membranes, albeit with much simpler composition. Then we reconstituted GluRs and found that the receptors preferentially insert into the HD comparing to the LD as shown in Fig. 2 [2]. This result demonstrates that bilayer thickness is a significant factor in the membrane self-assembly process. This can be expected to reveal signalling mechanism in synapses.
This research was supported in part by Bio-nanotechnology IRC in UK and
by the Strategic International Cooperative Program, Japan Science and Technology
Agency (JST).
[1] J. A. Allen et al., Nat. Rev. Neurosci. 8 (2007) 128.
[2] C. S. Ramanujan et al., 52nd Biophys. Soc. Meeting Abst. (2008) B363.
 |
 |
|||||
|
|